Tidyverse First Introduction
Nov 20, 2019
10 mins read
Contents (for MB2, 19/11/08)
- Tidyverse
- tidyr
- dplyr
- Excersice (data manipulation using
tidyranddplyr)
Tidyverse
Tidyverse has three meanings:
- Concept:
to use “tidy data” and to make the design of the tools used for data analyze more tidy. - Packages:
to manipulate data handling or data visualizing based on tidyverse (concept). - Organization:
is managingtidyverse(packages).
Today I explain about tidyverse (packages) mainly.
Recently, tidyverse, which became one of the dialects in R languages, is a powerful and useful collection of packages.
Many codes on the web are written using tidyverse.
So, if you want to learn R on the web, there is no less to know tidyverse.
I strongly recommend to read and demonstrate “R for Data Science (R4DS)” for studying tidyverse or data handling skills in R.
But this book has so many contents so that focus on some of them at this lecture.
First of all, to load the tidyverse and make it available in your current R session.
install.packages("tidyverse") #if you have not installed tidyverse
library(tidyverse)
Core tidyverse
NOTE: The following list is just only my thoughts.
- tidyr (create tidy data)
- dplyr (data transformation)
- purrr (apply function)
- ggplot2 (data visualization)
- stringr (string manipulation)
- readr (data import)
Today, I will explain only about tidyr and dplyr.
If you want to learn about other packages in tidyverse, please refer to RStudio Cheat Sheets or R for Data Science or Heavy Watal (jp lang).
tidyr
Almost packages in tidyverse operate based on “tidy data”.
Not only that, we can understand its meanings simply using “tidy data.”
But in the real world, we often encounter “non-tidy data”.
The tidyr package allows us to create “tidy data” easily.
What is “tidy data”?
“tidy”: arranged neatly and in order.
Hadley Wickham defined “tidy data” as follows.
In tidy data:
- Each variable forms a column.
- Each observation forms a row.
- Each type of observational unit forms a table.
Referrence: Tidy Data

by R4DS
Examples of “tidy data” and “non-tidy data”
“Tidy data” is like this:
table1
#> # A tibble: 6 x 4
#> country year cases population
#> <chr> <int> <int> <int>
#> 1 Afghanistan 1999 745 19987071
#> 2 Afghanistan 2000 2666 20595360
#> 3 Brazil 1999 37737 172006362
#> 4 Brazil 2000 80488 174504898
#> 5 China 1999 212258 1272915272
#> 6 China 2000 213766 1280428583
Each column means variables, and each row represents observations.
On the other hands, “non-tidy data” are like these:
table2
#> # A tibble: 12 x 4
#> country year type count
#> <chr> <int> <chr> <int>
#> 1 Afghanistan 1999 cases 745
#> 2 Afghanistan 1999 population 19987071
#> 3 Afghanistan 2000 cases 2666
#> 4 Afghanistan 2000 population 20595360
#> 5 Brazil 1999 cases 37737
#> 6 Brazil 1999 population 172006362
#> # … with 6 more rows
Because two types (cases and population) are common to country column and year column, table2 is “non-tidy data”.
table3
#> # A tibble: 6 x 3
#> country year rate
#> * <chr> <int> <chr>
#> 1 Afghanistan 1999 745/19987071
#> 2 Afghanistan 2000 2666/20595360
#> 3 Brazil 1999 37737/172006362
#> 4 Brazil 2000 80488/174504898
#> 5 China 1999 212258/1272915272
#> 6 China 2000 213766/1280428583
Because rate column consists of denominator and numerator, table3 is also “non-tidy data”.
Functions of tidyr
gather
table4a
#> # A tibble: 3 x 3
#> country `1999` `2000`
#> * <chr> <int> <int>
#> 1 Afghanistan 745 2666
#> 2 Brazil 37737 80488
#> 3 China 212258 213766
table4a contains year (1999 or 2000), but each year are columns. “gather” function takes multiple columns and collapses into key-value pairs, duplicating all other columns as needed.
gather(table4a, `1999`, `2000`, key = "year", value = "cases")
#> # A tibble: 6 x 3
#> country year cases
#> <chr> <chr> <int>
#> 1 Afghanistan 1999 745
#> 2 Brazil 1999 37737
#> 3 China 1999 212258
#> 4 Afghanistan 2000 2666
#> 5 Brazil 2000 80488
#> 6 China 2000 213766
Above scripts changed table4a to “tidy data”.
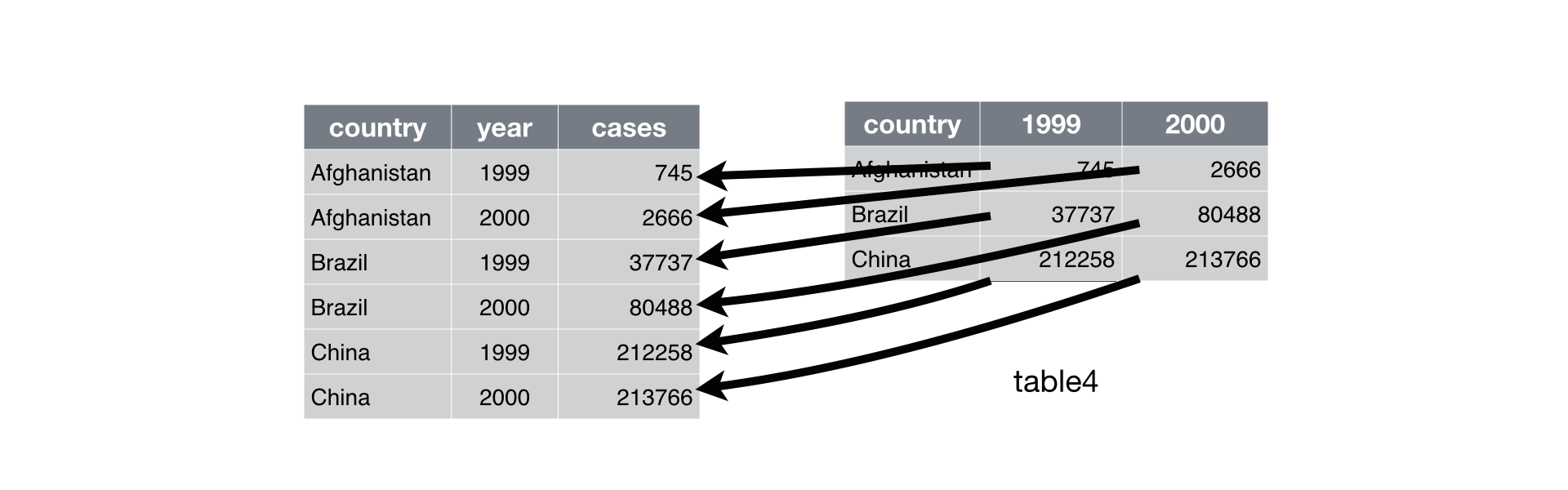 by [R4DS](https://r4ds.had.co.nz/)
by [R4DS](https://r4ds.had.co.nz/)
spread
spread function is the opposite of gathering.
table2 have observations which are scattered across multiple rows.
spread(table2, key = type, value = count)
#> # A tibble: 6 x 4
#> country year cases population
#> <chr> <int> <int> <int>
#> 1 Afghanistan 1999 745 19987071
#> 2 Afghanistan 2000 2666 20595360
#> 3 Brazil 1999 37737 172006362
#> 4 Brazil 2000 80488 174504898
#> 5 China 1999 212258 1272915272
#> 6 China 2000 213766 1280428583
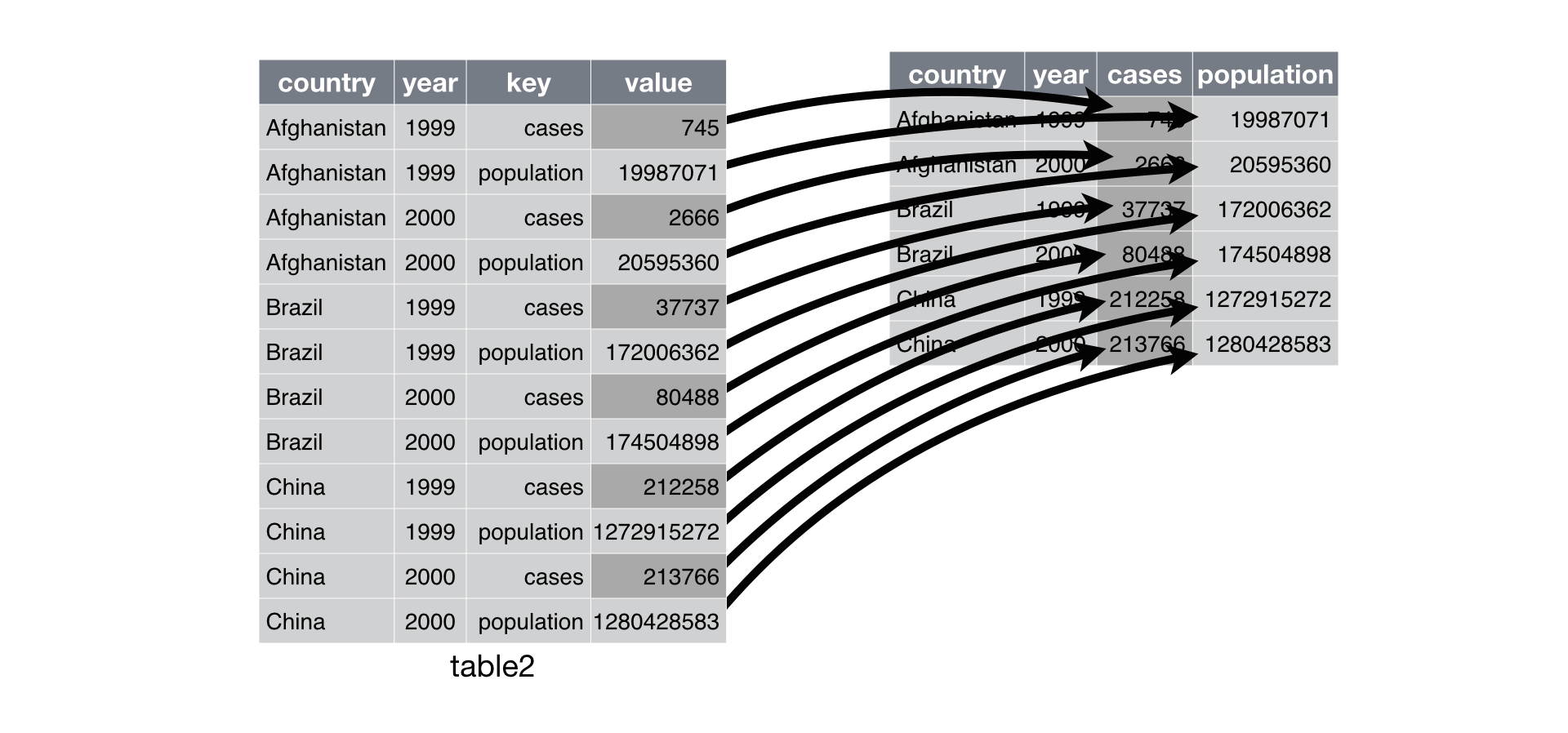
by R4DS
The tidyr package has more functions but the above lecture only introduces to spread and gather due to time.
More functions are here. Or we can confirm functions by running following commands.
help(package = "tidyr")
dplyr
dplyr is a powerful tool to transform data, like summarising, grouping, manipulating cases, manipulating variables, even combination.
pipe operator
%>% will forward a value, or the result of an expression, into the next function call/expression.
Actually, the pipe operator is function from magrittr package which is one of tidyverse.
The pipe operator makes R scripts more simple, readable, and hence easy to edit. This is because nested functions (the first script) are difficult to read and also lots of variables (the second script) confuse us.
Following two scripts will return same results.
spread(table2, key = type, value = count)
or
table2 %>%
spread(key = type, value = count)
%>% operator would demonstrate its power when applying many functions in one process.
Following three scripts also return the same result.
mutate(spread(filter(table2, country=="Afghanistan"),key = type, value = count), ratio=population/cases)
# `mutate` function will be discribed later.
or
Af <- filter(table2, country=="Afghanistan")
spreadAf <- spread(Af, key = type, value = count)
mutate(spreadAf, ratio=population/cases)
but you can utilize pipe operator,
table2 %>%
filter(country=="Afghanistan") %>%
spread(key = type, value = count) %>%
mutate(ratio=population/cases)
The first two are traditional R scripts. On the other hand, the third is a modern R script.
I strongly recommend using the pipe operator to make your code simple and readable. And I write all scripts using the pipe operator from here on.
functions of dplyr
I will explain only some of the functions because dplyr also has so many useful functions.
mutate
mutate function adds new variables, as used in the above scripts.
table1 %>%
mutate(ratio=population/cases)
Of course, mutate function can add columns using another dataset.
table1 %>%
mutate(rate=table3$rate)
In dialect tidyverse, it is common to access the columns as follows:
table1 %>%
mutate(rate=table3 %>% pull(rate))
Like above, %>% is also valid when passing as arguments.
pull command access the variables in the data frame.
mutate function is so useful but we must pass data of the same length as the original data frame.
If based on the tidyverse concept, we should operate at variables (columns) level of the data frame as possible.
So, we will often use mutate function.
The following figure is for illustrative purposes.

select
The select function filters the column levels.
If you want to do as same as table1$year, you should use pull command which is one of the commands in dplyr.
The pull command or $ operator return the data.frameRowLabels type.
But select function returns the data.frame type which columns name remains.
table1 %>%
select(countey)
The following figure is for illustrative purposes.

filter
The filter function filters the row levels.
filter function returns matched row when matching arguments.
table1 %>%
filter(country=="Afghanistan")
The above commands return rows including "Afghanistan" in country column.
Of course, you can also select by row numbers as below.
But if so, you have to use slice function.
table1 %>%
slice(1)
The following figure is for illustrative purposes.

inner_join
The _join function group (left_, right_, inner_ full_) combine two data.
The easiest to use function is inner_join which combine and returns only the rows that match the column specified by by.
table2 %>%
inner_join(table3, by=c("country", "year"))
The above commands returns the data combined with country and year columns.
The following figure is for illustrative purposes.
x has a, b, and c in A column and t, u, and v in B column.
On the other hand, y has the same variable as x like a and b in A column and t and u in B column.
In this case, inner_join combine as below:


group_by
group_by function makes groups.
This function is compatible with summarize function.
summarize
The summarize function summarize variables and create a new table of summary statistics.
mtcars %>%
as_tibble() # tibble is easy to read and use class because we can confirm all variable's names, numbers of rows and columns, and columns class.
#> # A tibble: 32 x 11
#> mpg cyl disp hp drat wt qsec vs am gear carb
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 21 6 160 110 3.9 2.62 16.5 0 1 4 4
#> 2 21 6 160 110 3.9 2.88 17.0 0 1 4 4
#> 3 22.8 4 108 93 3.85 2.32 18.6 1 1 4 1
#> 4 21.4 6 258 110 3.08 3.22 19.4 1 0 3 1
#> 5 18.7 8 360 175 3.15 3.44 17.0 0 0 3 2
#> 6 18.1 6 225 105 2.76 3.46 20.2 1 0 3 1
#> 7 14.3 8 360 245 3.21 3.57 15.8 0 0 3 4
#> 8 24.4 4 147. 62 3.69 3.19 20 1 0 4 2
#> 9 22.8 4 141. 95 3.92 3.15 22.9 1 0 4 2
#>10 19.2 6 168. 123 3.92 3.44 18.3 1 0 4 4
#># … with 22 more rows
mtcars %>%
group_by(cyl) %>% # grouping by `cyl` levels, which are 4, 6, or 8.
summarise(avg = mean(mpg)) # summarizing by the group (is `cyl` levels), and creation the averages column.
#># A tibble: 3 x 2
#> cyl avg
#> <dbl> <dbl>
#>1 4 26.7
#>2 6 19.7
#>3 8 15.1
The following figure is for illustrative purposes.

Exercise
Let’s enjoy data handling with tidyverse !!